For years, the ability to adapt to changing conditions has been seen as a hallmark of complex life forms. Now, scientists have engineered a system that mimics this adaptability in a test tube, blurring the lines between biology and computer science.
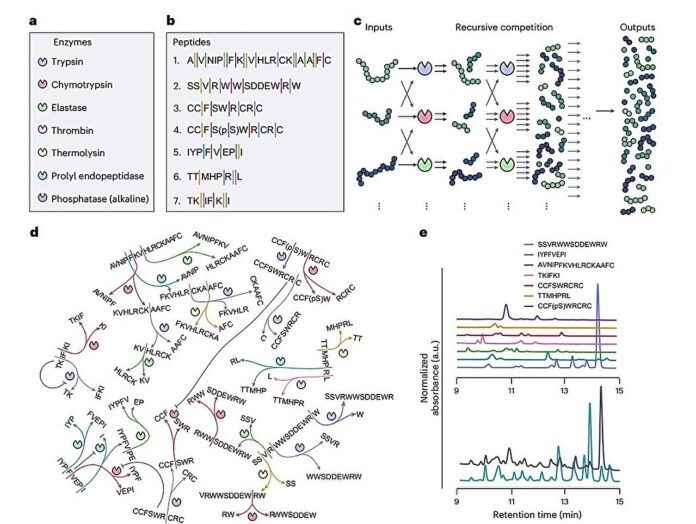
Imagine a bustling chemical factory where different molecules constantly vie for resources. This is essentially how researchers at institutions in the Netherlands and Australia built a decision-making “computer” from scratch. They constructed a network of enzymes called proteases, each vying with others for access to special peptides. These interactions weren’t pre-programmed; instead, the system self-organizes based on incoming signals, much like a biological cell reacting to its surroundings.
This chemical cocktail can do more than just react—it learns. The researchers demonstrated this by showing that their system accurately senses temperature changes within a range of 25°C to 55°C with impressive precision (about 1.3°C).
Mimicking Life’s Complexity
Living things are masters at gathering and processing information from the environment—whether it’s sensing nutrients, detecting light shifts, or feeling temperature fluctuations. This remarkable feat isn’t achieved through magic; intricate networks of molecules constantly communicate and react within cells.
Scientists have long been fascinated by these “network motifs”— repeating patterns found in natural chemical networks. They’ve used these motifs as blueprints to build artificial systems that mimic some aspects of biological information processing. But replicating the full complexity of living organisms has been elusive until now.
Recursive Interactions: The Key to Adaptability
The breakthrough lies in incorporating something called “recursive interactions”—where a reaction’s output becomes part of the input, creating a loop of continuous change and adaptation. Think of it like a message being sent back and forth, each time evolving slightly based on the previous exchange. This intricate feedback mechanism allows for a vast range of outputs from relatively simple starting points.
The researchers achieved this in their new system by building a complex network of seven enzymes and seven peptides. These peptides constantly compete for access to the enzymes, getting chopped up and reassembled in various combinations. The result is a dynamic, ever-shifting chemical landscape where the mixture of molecules changes dramatically depending on initial conditions like temperature or peptide concentration.
From Molecules to Decisions
This constantly changing molecular soup is analyzed in real time using a mass spectrometer—a tool that can identify individual molecules within a complex mixture. This data feeds into a simple algorithm that decodes these patterns and translates them into meaningful outputs, such as temperature readings, recognizing light pulses, or even detecting the passage of time.
This “chemical computer” could pave the way for smarter biosensors capable of responding to specific environmental cues in real-time. Imagine sensors able to detect subtle shifts in pH levels within a body, helping diagnose diseases earlier, or materials that change color based on temperature fluctuations, providing intuitive feedback in smart homes.
While still in its early stages, this research demonstrates the potential for engineering complex systems that learn and adapt from their surroundings—a remarkable feat blurring the lines between artificial intelligence and nature’s own ingenuity.